重要研究成果
第 1 頁
第 2 頁
林育如 博士
(2026)
Phys. Rev. Lett. 135, 093401 (2025).
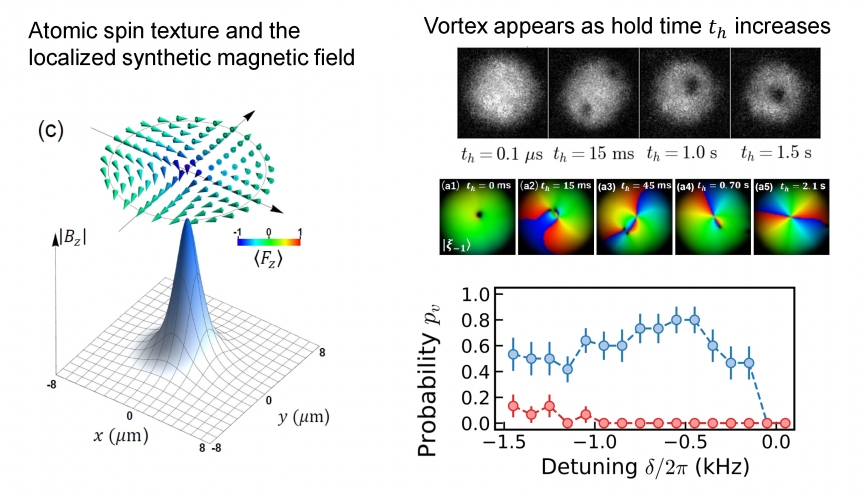
Gauge fields are ubiquitous in modern quantum physics. In superfluids, quantized vortices can be induced by gauge fields. Here we demonstrate the first experimental observation of vortex nucleations in light-dressed spinor Bose-Einstein condensates under radially-localized synthetic magnetic fields. The light-induced spin-orbital-angular-momentum coupling creates azimuthal gauge potentials $\vec{A}$ for the lowest-energy spinor branch dressed eigenstate. The observation of the atomic wave function in the lowest-energy dressed eigenstate reveals that vortices nucleate from the cloud center of a vortex-free state with canonical momentum $\vec{p} = 0$. This is because a large circulating azimuthal velocity field ~$\vec{p}-\vec{A}$ at the condensate center results in a dynamically unstable localized excitation that initiates vortex nucleations. Our observation has reasonable agreement with the time-dependent Gross-Pitaevskii simulations.
任祥華 博士, 陳應誠 博士, 肖恩 博士
(2026)
Phys. Rev. Lett. 136, 050802 (2026).
The so-called state-carving protocol generates high-fidelity entangled states at an atom-cavity interface without requiring high cavity cooperativity. However, this protocol is limited to 50% efficiency, which restricts its applicability. We propose a simple modification to the state-carving protocol to achieve efficient entanglement generation, with unit probability in principle. Unlike previous two-photon schemes, ours employs only one photon which interacts with the atoms twice - avoiding separate photon detections which causes irrecoverable probability loss. We present a detailed description and performance evaluation of our protocol under non-ideal conditions. High fidelity of 0.999 can be achieved with cavity cooperativity of only 34. Efficient state-carving paves the way for large-scale entanglement generation at cavity-interfaces for modular quantum computing, quantum repeaters and creating arbitrary shaped atomic graph states, essential for one-way quantum computing.
任祥華 博士, 陳應誠 博士
(2025)
Optica Quantum 3, 590 (2025).

Academia, governments, and industry around the world are on a quest to build long-distance quantum communication networks for a future quantum internet. Using air and fiber channels, quantum communication quickly faced the daunting challenge of exponential photon loss with distance. Quantum repeaters were invented to solve the loss problem by probabilistically establishing entanglement over short distances and using quantum memories to synchronize the teleportation of such entanglement to long distances. However, due to imperfections and complexities of quantum memories, ground-based proof-of-concept repeater demonstrations have yet been restricted to metropolitan-scale distances. In contrast, direct photon transmission from satellites through empty space faces almost no exponential absorption loss and only quadratic beam divergence loss. A single satellite successfully distributed entanglement over more than 1,200 km. It is becoming increasingly clear that quantum communication over large intercontinental distances (e.g., 4,000–20,000 km) will likely employ a satellite-based architecture. This could involve quantum memories and repeater protocols in satellites, or memory-less satellite-chains through which photons are simply reflected, or some combination thereof. Rapid advancements in the space launch and classical satellite communications industry provide a strong tailwind for satellite quantum communication, promising economical and easier deployment of quantum communication satellites.
張煥正 博士
(2025)
Adv. Funct. Mater. 36, e13406 (2026).

Nanoscale quantum sensing is playing an increasingly critical role across diverse areas of research, particularly in the rapidly evolving field of semiconductor nanoelectronics. In this work, ultrathin fluorescent nanodiamond (FND) films are developed to function as quantum sensors for in operando measurements of magnetic fields and temperature in semiconductor devices. FNDs are electrically insulating carbon nanomaterials containing nitrogen-vacancy (NV) centers, renowned for their exceptional photostability and distinctive quantum properties. An electrospray deposition method is first established to produce uniform, near-monolayer FND films on bipolar junction transistors (BJTs) and field-effect transistors (FETs) without compromising their performance. Then, optically detected magnetic resonance (ODMR)s is employed to detect magnetic fields and monitor temperature increases as electrical currents are passed through the FND-coated semiconductor chips. Finally, we introduce a technique called FND-based lock-in photoluminescence (PL) thermography, which enables wide-field, real-time temperature sensing and imaging of these actively operating BJTs and FETs with nanometric spatial and millisecond temporal resolution. In comparison to ODMR, this innovative PL thermography method offers enhanced practicality and ease of implementation, making it well-suited for diagnostic applications in semiconductor devices.
許良彥 博士
(2025)
J. Phys. Chem. Lett., 16, 1604−1619 (2025).
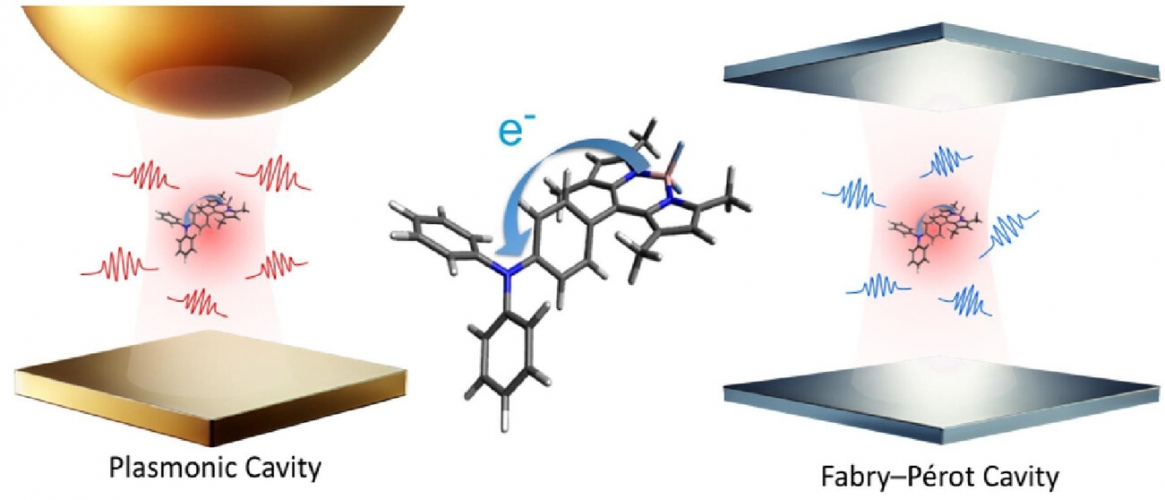
The interaction between light and molecules under quantum electrodynamics (QED) has long been less emphasized in physical chemistry, as semiclassical theories have dominated due to their relative simplicity. Recent experimental advances in polariton chemistry highlight the need for a theoretical framework that transcends traditional cavity QED and molecular QED models. Macroscopic QED is presented as a unified framework that seamlessly incorporates infinite photonic modes and dielectric environments, enabling applications to systems involving plasmon polaritons and cavity photons. This Perspective demonstrates the applicability of macroscopic QED to chemical phenomena through breakthroughs in molecular fluorescence, resonance energy transfer, and electron transfer. The macroscopic QED framework not only resolves the limitations of classical theories in physical chemistry but also achieves parameter-free predictions of experimental results, bridging quantum optics and material science. By addressing theoretical bottlenecks and unveiling new mechanisms, macroscopic QED establishes itself as an indispensable tool for studying QED effects on chemical systems.
目前位置:關於本所 / 重要研究成果 / 第 1 頁
中央研究院 原子與分子科學研究所版權所有 |
使用者條款、資訊安全與隱私權政策 |
保有個資檔案公開項目彙整表 |
行動版
地址: 106319 台北市羅斯福路四段一號 或 106923 臺北臺大郵局 第23-166號信箱
電話:886-2-2362-0212 傳真:886-2-2362-0200 電子郵件:iamspublic@gate.sinica.edu.tw
最後更新於 2026-03-09 19:05:23
地址: 106319 台北市羅斯福路四段一號 或 106923 臺北臺大郵局 第23-166號信箱
電話:886-2-2362-0212 傳真:886-2-2362-0200 電子郵件:iamspublic@gate.sinica.edu.tw
最後更新於 2026-03-09 19:05:23
 中央研究院 原子與分子科學研究所
中央研究院 原子與分子科學研究所
